Tag: chromatin
There is a foundation of transcription for human cell types
by admin
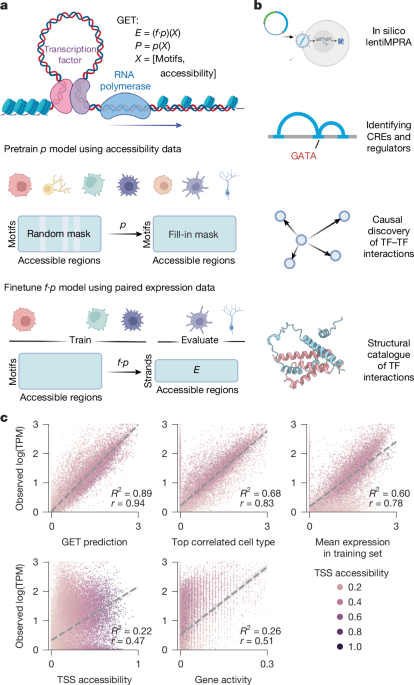
A model was used to learn regulatory grammar in the GET architecture using chromatin accessibility data from a variety of human cell types. The model was trained on the fetal accessibility and expression atlas and was able to complete in 8 h. It is possible to perform large-scale screening with the help of inference for genes in a single cell type.
Read MoreBrain-wide correspondence ofgenomics and projections
by admin
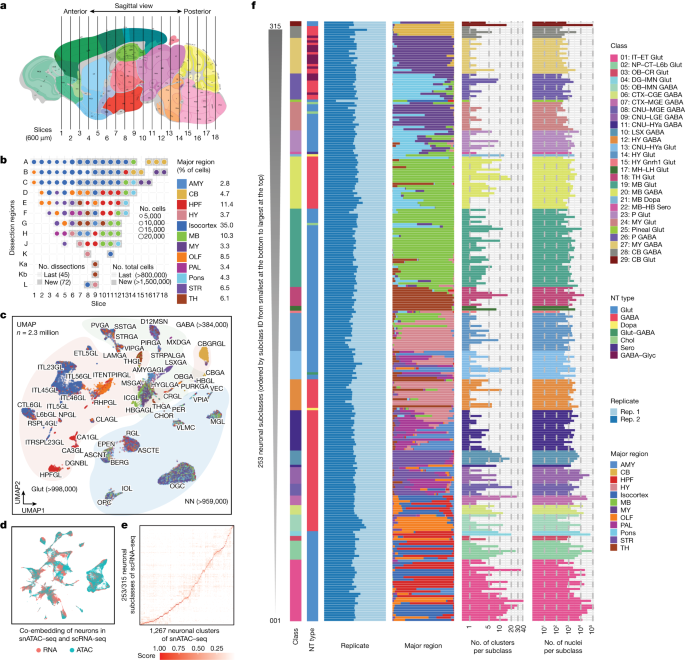
Our study found that there’s statistically significant higher enrichment (P = 0.004) of chromatin interaction signals at the corresponding-specific cCRE-gene pair anchors, compared with non-corresponding pair anchors. This suggests that cCRE-gene pairs are more likely to interact in cell types in which the cCREs are active. We used snRNA-seq cells for GWAS with MAGMA v.1.10 to compare human and mouse gene sets.
Read More