A coronaviruses assembly drug that targets the viral vesicles
by admin
Statistical study of the left lung of Syrian golden hamsters infected with SARS-CoV-2 and vehicle or JNJ-9676
The project 062/2020 was approved by the ethics committee of KU Leuven. The hamster infection model of SARS-CoV-2 has been described previously72. 5 animals in each group were needed for statistical significance in the Syrian golden hamster studies. After arriving, the animals were randomly assigned to groups. No blinding was performed during the experiment. Female hamsters (Janvier Laboratories), 8–10 weeks old, were inoculated intranasally with 50 μl containing 2 × 106 TCID50 SARS-CoV-2 B1 (day 0). The animals were treated according to the schedule with the vehicle or JNJ-9676. Animals were dosed BID at 08:00 and 16:00. Viral RNA and infectious virus levels in the right lung were quantified using RT–qPCR and end-point virus titration, whereas left-lung samples were subjected to histopathological scoring, as described previously72 (Fig. 3b–e).
In the photo. 3e, representative images are shown of H&E-stained left lung lobes of Syrian golden hamsters infected with SARS-CoV-2 and vehicle or JNJ-9676 treated. A full cross section of the left lung of each group of animals was assessed by a skilled pathologist.
In Vitro and In Viral Studies of the Synthesis of JNJ-9676 / Nirmatrelvisa and Molnupiravar in the Presence of SARS-CoV-2
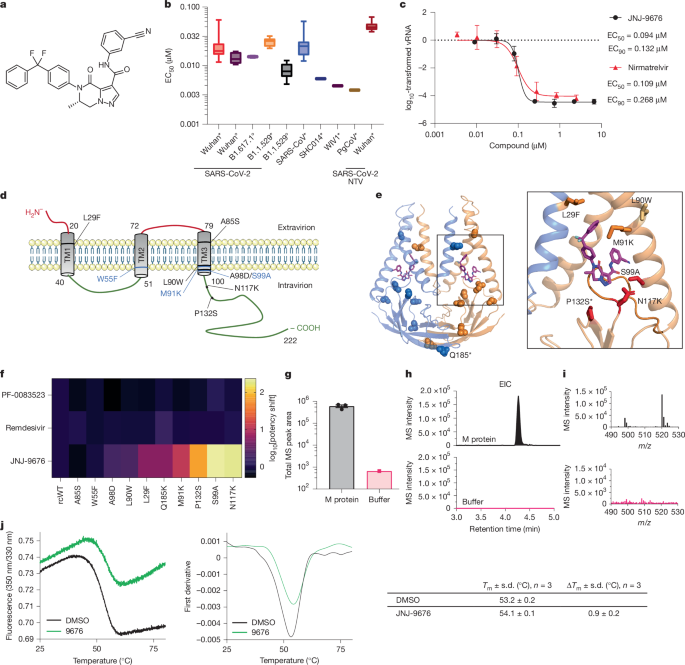
In Extended Data Fig. 2g, uncut western blots are shown of purified M proteins. These blots were generated once as a quality control of the protein obtained.
In a data diagram. A micrograph from the data collection is shown. There were 12,988 images taken to get this image.
The synthesis of JNJ-9676 is described in patent WO-2024/008909 and in the Supplementary Methods. Molnupiravir was ordered at MedChemExpress (HY-135853) and nirmatrelvir was synthesized according to literature procedures59. JNJ-9676, molnupiravar or nirmatrelvisa were dissolved in 100 mM of dimethyl sulfoxide for in vitro experiments. For in vivo experiments, JNJ-9676 was dissolved in 100% polyethylene glycol 400 (PEG400) as stocks of 75, 25 or 8.33 mg ml−1, molnupiravir was dissolved in 100% PEG as a stock of 300 mg ml−1 and nirmatrelvir as a stock of 250 mg ml−1.
JNJ-9676 or vehicle (0.2% DMSO) was prepared in MucilAIR medium (Epithelix) and added on day 1 to the basal compartment of 24-well MucilAIR plates (Corning Costar clear PS plates, Merck) with pooled nasal epithelial cells. Then, 1 h later, the inserts were infected with SARS-CoV-2 (multiplicity of infection (MOI) of 0.1, B1 strain) for 1 h at 37 °C followed by three PBS washes. At 24 and 48 h, apical washes were collected. The cells were lysed using 200 l of RLT buffer. The MagNA Pure 96 DNA and Viral NA Small Volume Kit was used for the apical wash and the MagNA Pure 96 Cellular siRNA Kit was used for the cell extracts. For the apical washes, an external lysis step (Roche lysis buffer) was included before the RNA extraction. One-step reverse transcription quantitative polymerase chain reaction (RT–qPCR) was performed on extracts using the LightCycler Multiplex RNA Virus Master kit (Roche) and SARS-CoV-2 primers and probe (located in nucleocapsid gene; https://stacks.cdc.gov/view/cdc/84525) and in-house designed β-actin primers and probes. A real-time PCR instrument was used to calculate absolute quantites using a series of dilutions of the ‘Scarps- CoV-2’ nucleic acid fragments.
Cells wereseeded the day before the test to make sure they were prepared for the test. The compound dilutions were mixed with virus and put on cells for 30 minutes at room temperature. The cells were then put into a solution of the relevantviruses at 37 C or 32 C. Cells were washed twice with PBS, the first wash was followed by the compound containing medium and the second was used to further acclimatize the cells to 37 C. Three-fold serial dilutions of compounds in the H CoV- 229E test were made in 96-well plates. After that, Huh7 cell suspension was added to assay wells at a density of 10,000 cells per well and cells were infected with HCoV-229E at an MOI of 0.01. The cell viability was determined after 5 days. The VeroE6 cells had to be seed in 96-well plates one day before the risk of the disease. On the following day, a couple of serial dilutions of compound in infection medium with 0.25 M. The cells were added with a p-GP Inhibitor, and the rest of the plate was exposed to either a strain of the disease or a different strain. The other half of the plate was mock infected to determine the possible cytotoxic effect of the compounds in parallel. The CellTiter 96 kit was used to measure the cell viability of the cells after they had been affected by the disease. The values of the wells were normalized to the average signal of the cells who didn’t get sick. The EC50 and CC50 values were determined by nonlinear regression.
The viruses, derived from infectious clones of Pg CoV, were isolated and made into working stock.
Measurement of toxicity of recombinant SARS-CoV-2 M and 100 M of JNJ-9676 for infection using HisTrap cells (Invitrogen)
The analysis of toxicity was performed on day 5 in cells that had been treated but didn’t have any infections.
The toxicity was assessed by exposing insert to the same concentration of JNJ-9676 as it would be if it were treated for a drug allergy. Brefeldin (0.3 µM; internally synthesized) was used as a toxicity control.
Cells were trypsinized and resuspended in PBS, after which they were freeze-thawed 3× on liquid nitrogen to release intracellular viral particles. Cellular debris was removed by centrifugation (5 min at 13,400 rpm and 4 °C) and 50 µl of the supernatant was used to determine the infectious titre by plaque assay.
pcDNA3.4 vectors containing FabB heavy chain and light chain were co-transfected into Expi293F cells (Invitrogen) according to the manufacturer’s protocol and incubated for 96 h at 37 °C with 8% CO2.
A 10 liters of medium was loaded onto it. The HisTrap column has a flow rate of 8 liters per minute. The column was washed with 6 CV of wash buffer (20 mM sodium phosphate pH 6.5, 150 mM NaCl, 20 mM imidazole) and eluted with over 5 CV using a 39.2–500 mM imidazole gradient prepared in buffer (20 mM sodium phosphate pH 6.5, 150 mM 500 mM imidazole, is NaCl. FabB peak fractions were then put onto a HiLoad 16/600 Superdex 75 pg column with a buffer of 20 mM. NaCl).
Experiments were performed in a total volume of 10 µl. Using a Prometheus NT.Plex instrument was used to measure the melting temperatures. The samples were prepared in a 384-well plate with 0.5 mg ml−1 purified recombinant SARS-CoV-2 M and 100 µM of JNJ-9676 in 20 mM HEPES pH 7.5, 150 mM NaCl, 0.001% LMNG (w/v), 0.0001% CHS (w/v), 0.00033% GDN (w/v) and 1% DMSO (v/v). The samples loaded into standard‐grade glass capillaries were measured under a temperature range of 25–95 °C with a temperature gradient of 1 °C min−1, and the intrinsic protein fluorescence at 330 and 350 nm was recorded. The data were analysed using PR.ThermControl v.2.1.6 (NanoTemper Technologies) (technical replicates ≥ 3).
The offline ASMS experiment consisted of the preparation of three sample types: compound QC, protein target (M protein) and no-protein control (breakthrough).
For the preparation of SEC filter plates for offline ASMS, 130 µl of pre swollen Bio-Gel P10 resin slurry was added to each well of a low-protein-binding Millipore HTS 384 HV filter plate (hereafter, size-exclusion plate) with a 0.45 µm Durapore (PVDF) membrane (MZHCN0W10). The size-exclusion plate was placed into a 4 °C refrigerated centrifuge, centrifuged at 1,000g for 2 min and the flowthrough was discarded. The four washes were done using 50 l buffer with 20 mM HEPES and a pH 7.5. NaCl, 0.001% LMNG, 0.0001% CHS, 0.00033% GDN and 2% DMSO, whereby the flowthrough from each wash was discarded after centrifugation at 1,000g for 2 min. A long aliquot of 20 t of 5 mM compound was transferred to the four separate wells of a 384-well, natural V- bottom plate from the source plate using an acoustic liquid handler. The sample of the solution was dilution using a buffer to obtain a concentration of M and 2% DMSO. Then, 20 µl of the resulting working protein stock was dispensed into three wells containing compound to yield a final concentration of 5 µM (3 technical replicates). A separate working stock was prepared without protein and placed into the remaining compound well to control for compound breakthrough. The plate was centrifuged at 1,000g for 1 min at room temperature and incubated at 25 °C for 30 min.
The samples were swiftly loaded into the size-exclusion plate, which was emptied at a rate of 1,000g per minute in 2 minutes to minimize compound breakthrough. The flowthrough was sieved out with 15 lMS-grade water and thencentrifugationd for 5 minutes at a room temperature to remove any insoluble particles.
The compound was transferred from the source plate into a 395-wellplate, and combined with 25% acetonitrile in a 2% DMSO solution.
All liquid chromatography–mass spectrometry (LC–MS) analyses were performed on an Agilent 1290 Infinity II uHPLC system coupled to an Agilent 6545XT qTOF using the Agilent MassHunter (v.10.0) software. A 4 µl sample injection was loaded with water as a loading solvent onto the reversed-phase column (2.1 × 35 mm ACQUITY UPLC BEH C18 column, 130 Å, 1.7 µm), heated to 40 °C. The mobile phases consisted of water and acetonitrile and contained a small amount of formic acid. The method uses a constant flow rate of 0.1 ml min1 and consists of a 1 min wash with 5% solvent B, followed by a shallow slope from 5% to 85% B over 1.9 min and a hold for 1 min. The MS instrument was operated in positive polarity mode with centroided data acquisition, where the source was set to a 350 °C drying gas temperature and 13 l min−1 drying gas flow rate; 375 °C sheath gas temperature and 12 l min−1 sheath gas flow rate; capillary voltage of 3,300 V; nozzle voltage of 500 V; nebulizer pressure of 50 psi; fragmentor of 125 V; and skimmer of 50 V. A reference mass solution consisting of purine and HP-0921 (Agilent, G1969-85001) was prepared according to the manufacturer’s instructions and infused to apply automatic mass correction to all spectra acquired from 110 to 1,100 m/z at a rate of 1 spectrum per second.
The data was processed using an error tolerance window of 3 ppb, where the M+H,M+Na, andM+K mass were merged.
Real-time cryo-EM data collection and analysis at the PELCO easiGlow discharge with a vitrrobot
The glow discharge was subjected to by the PELCO easiGlow discharge cleaning system. The 3 l recombinant M product sample was used to apply to the EM grids and was then cultured with a vitrrobot. The freeze in liquid ethane was done with liquid nitrogen. Data collection on the Glacios microscope was done using a software program. Micrographs were taken at ×105,000 magnification using a Facon4 detector (Gatan) in counting mode. 40 frames with a total dose of 40 e 2 was recorded for each 6 s exposure. The digital micrographs had a physical Size of 0.910.
cryo-EM data collection was monitored in real time using a live software. Image preprocessing steps, including patch motion correction, patch contrast transfer function (CTF) estimation, blob particle picking (100–200 Å diameter) and extraction, were performed simultaneously. A total of 12,988 raw micrographs was recorded during a 4-day data collection session using the Glacios microscope. Acceptable 2D classes served as templates for particle repicking. One round of live 2D image classification yielded approximately 1.2 million good particle images. The particles were used in the reconstruction. The first round of five starting 3D models were calculated, resulting in one major 3D class, followed by a second round of four 3D classes. One major class underwent non-uniform 3D refinement and local refinement using 484,610 particles and was further refined to a 3D EM map with an average resolution of 3.06 Å.
Resolutions were estimated by applying a soft mask around the protein complex density using the gold-standard (two halves of data refined independently) FSC = 0.143 criterion. Before visualization, all density maps were sharpened by applying different negative temperature factors along with the half maps and used for model building. Res Map was used to determine local resolution. Information about the processing of the data can be found in Extended Data Fig. 6a–f.
The Alphafold model62 of Fab–B was fitted to the M–Fab–B-CIM map using the UCSF Chimera Fit in map tool63. An initial round of molecular-dynamics flexible fitting was then done on the combined model using Namdinator64 and then manually adjusted in Coot65. The compound was added with the help of the ligand builder tool. When selecting the pose of CIM-834, the following were taken into account. First, the morphology of the density. The piperidine andpyrimidine rings were found in the density near the M two-fold symmetry axis. The density for the opposing end of the density was thinner and less well resolved (Extended Data Fig. 6), consistent with the increased flexibility of the molecule beyond the amide position. The positioning of hydrogen-bonding partners. The selected orientation of CIM-834 positions the pyridazine ring in proximity to S99 and N117, allowing for potential direct or water-mediated hydrogen bonds. The model was refined using iterative cycles of model building and real space refinement using eLBOW. Model validation was done with the help ofMolprobity68.
GraphPad Prism applied to the study of lung tissue stained with haematoxylin and eosin for histological examination
For histological examination, the fixed lung tissue sections (5 μm) were analysed after staining with haematoxylin and eosin and scored blindly for lung damage by an expert pathologist. The scored parameters, (cumulative score, 1 to 3), were as follows: congestion, intra-alveolar haemorrhagic, apoptotic bodies in the bronchus wall, necrotizing bronchiolitis, perivascular oedema, bronchopneumonia, perivascular inflammation, peribronchial inflammation and vasculitis.
GraphPad Prism has a statistical analysis tool called GraphPad Prism v.9.6.0 that is used for the analyses. The lung viral-load data was transformed using a log10 transformation. The one-way analysis of variance with dk’s multiplicity correction was used to estimate the mean differences between the treatment and vehicle groups.
The non-paradigm Kruskal–Wallis test by ranks was used if normality couldn’t be assumed in the outcome variable or lung histopathology. Multiple testing was taken into account when applying a post hoc test with the Benjamini-Hochberg correction. The significance level was used.
The graphs were prepared using the GraphPad Prism. Figures and schemes were created using BioRender.com and Adobe Illustrator 28.1 (Windows).
The graphs used in the study were in GraphPad Prism and the tests used in them were shown in the figure legends. The 3 independent preparations that produced 2D EMs were used to acquire the 2D DEMs. The majority of the samples were prepared in one preparation with the exception of the HPF samples.
Microsomal Stability Measurements Using a Biomek Automate and a Nanometer Nanoscale Robotized Greiner Plate for Determination of the IC50
Compound microsomal stability was determined using mouse microsomal fractions (Gibco; final protein concentration, 0.5 mg ml−1), with hamster microsomal fractions (Xenotech; final protein concentration, 0.5 mg ml−1) and with human microsomal fractions (Xenotech; final protein concentration, 0.5 mg ml−1) at a substrate concentration of 1 μM with or without NADPH (final concentration, 1 mM). Incubations were done at 37 °C for 120 min in duplicates. 25 l of the reaction mixture was taken and then quenched in 300 l of acetonitrile. The resulting suspension was vortexed and centrifuged, and the supernatant was diluted with water (two-fold). The half-life of the compound was determined with the analysis of the solution by liquid chromatography and tandem mass spectrometry.
The IC50 was determined using this equation: percentage of active enzyme = 100/(1 + I2/IC50), where I is the concentration of inhibitor and 100% of activity is the fluorescence intensity without the inhibitor. Curve fitting was used to determine the IC50. For each measurement, results were obtained in triplicate.
Reaction assays were stopped by adding 20 µl EDTA (100 mM). Positive and negative controls consisted of a reaction mix with 5% DMSO (final concentration) or EDTA (100 mM) instead of compounds, respectively. Reaction mixes were then transferred to a Greiner plate using a Biomek I5 automate (Beckman). PicoGreen fluorescent reagent was diluted to 1/800 in TE buffer, according to the manufacturer’s instructions, and 60 μl of reagent was distributed into each well of the Greiner plate. After 5 min in the dark at room temperature, the plate was placed in a Tecan Safire2 or ClarioStar and read at 480 and 530 nanometers.
Reactions were done in a volume of 40 µl on a 96-well Nunc plate. The experiments were robotized with the use of the BioMek I5 automate. Then, 2 µl of each diluted compound in 100% DMSO was added in wells to the chosen concentration (5% DMSO final concentration). For each assay, the (nsp8L7 + nsp8) mix was distributed in wells after an 8-min incubation at room temperature to pre-form the active complex. Nsp12 was put into wells and put to sleep for 8 minutes. Reactions were started by adding the UTP + poly(A) template mix and were incubated at 30 °C for 20 min, using 350 nM of poly(A) template and 750 µM of UTP final concentration.
In the IC50 buffer, there are seven different increasing concentrations of the compound, which led to 50% inhibition of the polymerase-mediated RNA synthesis. 76) in complex with 450 nM nsp8 and 450 nM nsp8L7 (ref. 77).
The activity and inhibition were determined by FRET using black 384 well HiBase non-binding plates. In brief, increasing concentrations of inhibitor were incubated with purified PLpro protein (55 nM) in the presence of 5 μM of a fluorescent synthetic peptide (Dabcyl-FTLKGG↓APTK-Edans, Genscript) in HEPES buffer (20 mM, pH 6.5) containing 120 mM NaCl, 0.4 mM There are four substances namely: dtt, eda, mM and glycerol. The final concentration of DMSO was adjusted to 0.5%. The fluorogenic peptide separates the Edans/Deabcyl fluorophore–quencher pair. The 40 min followed by the enzymatic reaction was done using a Tecan Safire2 FLURITATION, which showed the increase in emission at the marcos. Enzymatic activities were estimated by taking into account the slope of the curve and the activity that was measured in the absence of a drug.
The buffer contained 20 mM HEPES, 120 mM, and a fluorescent plate with the cleavage site of the Mpro. NaCl is 0.4 mM. EDTA, 4 mM DTT, 20% glycerol, pH 7.0, was used for the inhibition assay. The Edans generated as a result of Mpro’s cleavage of the substrates was observed at an emission wavelength of over 450 lm using a Tecan Spark multimode microplate. The compound was made into a stock solution with 100% DMSO. The IC50 of CIM-834 was determined after 10 minutes of reaction buffer at 37 C and 500 M of Mpro and/ or CIM-834 at different concentrations. At the end, the FRET substrate at a final concentration of 50 µM was added to each well at a final total volume of 100 µl to initiate the reaction. The IC50 value was calculated with the help of the GraphPad Prism 9.2.0 software. Data is presented as a mean sd. if the compound’s activity was measured in triplicates. Two positive control assays were also done against Mpro.
Reverse-engineered SARS‐CoV‐2 Wuhan with the M-protein P132S substitution was constructed by mutating nucleotide positions 26916–26918 from CCG to AGC in a bacterial artificial chromosome vector containing a full-length cDNA copy of the genome of SARS-CoV-2 strain SARS-CoV-2/human/NLD/Leiden-0008/2020 using the following primer pair for two-step en passant recombineering in Escherichia coli48:
There were 100mm dishes with 2 106 cells per dish. On the next day, cell-culture medium was changed to one with varying concentrations ofCIM-832 (0.0 to 8.0 M). Cells were co- transfected with two pieces of the M(P134S) gene and one piece of the E-hsV-encoding gene using TransIT-LT1 Transfection Reagent. Then, 48 h after transfection, VLPs and cell lysates were collected and stored as described previously54 for MERS-CoV VLPs. lysate and VLP samples were separated from the Laemmli loading buffer on the basis of polyacrylamide gel. A primary anti-V5 and secondary HRP-labelled goat anti-mouse antibody was used for the immunoblotting. Tubulin was used as a loading control and stained using a mouse anti-β-tubulin IgG1 antibody (Sigma-Aldrich, T5201, clone TUB 2.1, 1:2,000). The band quantification function was used to quantify the western blotting bands. Supplementary fig. 1 is a record of all unprocessed blots.
Interactions between M and CIM-834 were calculated using LigPlot71 and UCSF ChimeraX72. The mean square deviation was obtained using the matchmaker tool. Figures were generated using UCSF ChimeraX72, and structural-biology applications used in this project were compiled and configured by SBGrid73.
The final concentration of 44.47 M Mprotein, 100 m CIM-834 and Fab–E/B was achieved for the Fab–E and Fab–B complexes. The components were mixed with a buffer solution of 20 mM HePES-Na OH and 150 mM. Let’s say NaCl is0.00075% and 2 millionNG and 2 millionCHS are the same. 15 min was spent on ice to prepare the sample for vitrification. The sample was put into a pipette and then plunge-frozen in liquid ethane using theThermo Fisher Scientific’s Vitrobot mark. The Netherlands Center for Electron nanoscope collected the two datasets. The grid was loaded into a Titan Krios electron microscope, which was powered by 300 kv and had a K3/Gatan energy filter on top of it. Fab–E and Fab–B were done in super-resolution mode at a nominal magnification of 81,000 and 105,000, respectively. A total of 5,058 movies were recorded for the Fab-E and Fab-B complexes. Detailed data-acquisition parameters are summarized in Extended Data Table 2.
For tomography, semithin sections of 200 nm or 300 nm were screened and imaged using a Tecnai F30 microscope (Thermo Fisher Scientific) equipped with a Gatan OneView camera. Target positions were manually selected and acquired at 15,500 magnification. The tilt series were reconstructed using a computer program. The methods for segmenting selected tomograms were done manually with the help of the Amira-Avizo software.
Cells prepared on sapphires for high-pressure freezing were prefixed and exported from BSL3 in 6% PFA, similarly to the samples on coverslips. The samples were immersed with 100 mM of PHEM buffer. PHEM with 15% BSA as a cryo-protectant and high-pressure frozen using a BalTec HPM-010 with carriers forming a 40 µm-deep cavity (3-mm aluminium carriers, type B 0/0.3 mm and type 748 0.04/0.020 mm, Engineering Office M. Wohlwend). High-pressure frozen samples were then subjected to freeze substitution in a Leica AFS II using cryo-tubes with screw caps and rubber sealing rings, containing 1 ml of fixative cocktail composed of 0.2% OsO4, 1% uranyl acetate and 5% water in acetone. The AFS chamber temperature was increased over the course of 24 h as follows: 1 h at −90 °C; 8 h at −90 °C to −80 °C; 8 h at −80 °C to −50 °C; 2 h at −50 °C to −20 °C; 2 h at −20 °C to 0 °C. After being washed with dry acetone and ice, the samples were then PROCESSED using a microwave twice, at 250 mW and with increasing concentrations of Epon 912 in the samples. sapphires were transferred to plastic moulds and put in the freezer for 72 hours after the last 100% step. Ultrathin and semithin sections of 70 nm or 300 nm, respectively, were generated for both chemically fixed and high-pressure frozen samples using a UC7 Leica ultramicrotome and a 30° diamond knife (Diatome) and collected on Pioloform-coated slot grids. Grids were post-stained for 5 min with 3% uranyl acetate in 70% methanol and 2 min with lead citrate. To locate the infected cells, the Serial-EM Navigator functionality and a procedure adapted from ref. 55 were used to map the central section of a ribbon of 5 on a JEOL 1400 equipped with a TemCam-F416 camera. A map containing a group of cells was used to identify and store them, while a Matataki sCMOS camera was used to acquire montages over the whole perinuclear region of the cells.
Source: A coronavirus assembly inhibitor that targets the viral membrane protein
VeroE6+GFP and A549 ACE2+TMPRSS2 cells (African monkey kidney cell line with green fluorescent protein) in modified Eagle medium supplemented with 10% v/v heat-inactiv
The ethics committee of animal experimentation ofKU Leuven approved the housing conditions and procedures for the animals.
GS-441524 was obtained from MedChem Express (HY-103586). Cell Signaling Technology purchased Hydroxychloroquine. Nirmatrelvir (PF-07321332) was from Wuxi.
VeroE6–GFP cells (African monkey kidney cell line expressing green fluorescent protein; provided by M. van Loock, Janssen Pharmaceutica40) were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% v/v heat-inactivated fetal bovine serum (FBS) The geneticin is given in the form of 1 gmil. VeroE6–mCherry cells (generated as described in ref. The DMEM was supplemented with 10% of heat-inactivated FBS and 10 g ml1 blasticidin. The A549ACE2+TMPRSS2 cells (a human lung carcinoma cell line overexpressing human ACE2 and human TMPRSS2 receptors), used for antiviral studies, were from InvivoGen (a549d-cov2r, A549-Dual hACE2-TMPRSS2 cells) and were cultured in DMEM supplemented with 10% v/v heat-inactivated FBS, 300 μg ml−1 hygromycin, 0.5 μg ml−1 puromycin and 10 μg ml−1 blasticidin. In-house transduction was built from A549, obtained from ATCC, using pWPI and 500 g geneticin. These cells were supplemented with a range of antibiotics, including penicillin, streptomycin, and non-essential Amino acids. All assays using VeroE6–GFP, VeroE6–mCherry and A549ACE2+TMPRSS2 involving virus replication were performed in the respective cell growth medium containing 2% (instead of 10%) FBS. At 37 C and 5% CO2, all cell cultures were done. BHK-21 cells, obtained from the ATCC, were kept in Glasgow MEM (Invitrogen)” and were supplemented with 5% v/v FBS” and 100 U grams penicillin. For transfection experiments, cells were maintained in Eagle’s minimal essential medium (EMEM), as described42. Huh7 cells were cultured in DMEM supplemented with 10% FBS and 1 mM Glutamax.
Nirmatrelvir (from Excenen, batch EXA5024) was formulated as a 100 mg ml−1 and 33.3 mg ml−1 stock (for 300 mg per kg and 100 mg per kg dosing, respectively) in a vehicle containing 43% absolute ethanol and 27% propylene glycol (Sigma) in sterile distilled water. The total amount of components in CIM-832 was 20% propylene glycol (Sigma), 1% Tween 80, and 85% pH 5 citrate buffer. To evaluate in vivo efficacy, male SCID mice (CB-17/Icr-Prkdcscid/scid/Rj; Janvier Laboratories) 7–9 weeks old were treated by oral gavage with either the vehicle (n = 12, twice a day) or CIM-834 at 100 mg per kg (n = 12 twice a day and n = 12 once a day) or nirmatrelvir at 300 mg per kg (n = 12, twice a day) or 100 mg per kg (n = 6, twice a day), starting from day 0, just before infection with the beta variant B.1.351 (hCoV-19/Belgium/rega-1920/2021; EPI_ISL_896474, 2021-01-11). Animals were inoculated with 40l containing 105 TCID50 and anesthetized with isoflurane for virus infections. SARS-CoV-2 beta variant (day 0). In the therapeutic set-up, animals were infected on day 0 and treatment with CIM-834 (100 mg per kg, twice a day) was initiated 24 h, 30 h or 48 h after infection. Mice were housed in individually ventilated cages with three mice per cage and monitored daily for weight changes and any clinical signs. At day 3 after infection, animals were euthanized by intraperitoneal injection of 100 µl Dolethal (200 mg ml−1 sodium pentobarbital, Vétoquinol SA), and the lungs were collected. The infectious viral lung loads were quantified. To prevent carry-over of the compound during determination of infectious virus titres, the cells were washed and given fresh medium, immediately after incubation for 2 h with lung homogenates. Cells were subsequently incubated at 37 °C for three days before TCID50 read-out.
All plasmids were validated by Sanger sequencing (Macrogen). The use of a 3 ms interval BTX system was not used for preparation of viral DNA, except for the use of an ECM 830 Square Wave system. The cells were put in medium with 10% FCS. After 6 h at 37 C, the medium was removed and replaced by the new medium with only 0.11% FCS. Four days later the virus stocks were collected and subjected to whole- genome Sequence Determination with the help of the Vanmechelen Institute.
For this, 5-fold dilutions of cell lysate were added to VeroE6 cell monolayers in twelve-well plates for 1 h at 37 C. The inoculum mixture was replaced with 0.8% MSM in addition to 2% FBS. After three days of incubation at 37 °C, the overlays were removed, the cells were fixed with 3.7% PFA and stained with 0.5% crystal violet, and plaques were counted visually.
The images show H&E-stained left lung lobes of Syrian golden hamsters infected with SARS-CoV-2 and vehicle or JNJ-9676 treated. Cells were washed twice with PBS, the first wash was followed by the compound containing medium and the second was used to further the cells to 37 C. JNJ-9676 or vehicle was prepared in MucilAIR medium (Epithelix) and added on day 1 to the basal compartment of 24-well MucilAIR plates.