Clinical classification of the BRCA2 variant is based on genome editing
by admin
In Situ Machine Learning for Genome-wide Variant Effect Prediction in the Early Stages of BRCA2 Gene Expression: Detection of Pathogenousity and Neutrality with MutateX
In this case, deep learning techniques have been used to improve genome-wide variant effect prediction. In the next decade, the Genome Med. 13, 31 will be published.
Richardson, M. E. et al. Strong functional data for both pathogenicity and neutrality classify the variant as uncertain significance. Am. J. Genet. 108, 458–468 (2021).
Tiberti, M. et al. MutateX: an automated pipeline for in silico saturation mutagenesis of protein structures and structural ensembles. Brief. Bioinformatics 23, bbac074 (2022).
Source: Saturation genome editing-based clinical classification of BRCA2 variants
Automated interpretation of clinical variants in a genomic dataset by means of the VarSome API: a case study of BRCA1 and BRCA2
de Bruijn, I. et al. AACR Project GENIE biopharma Collaborative data can be analyzed and visualized. Cancer Res. 83, 3861–3867 (2023).
Sorrentino, E. et al. Automatic ACMG interpretation of clinical variants can be achieved by integration of VarSome API into an existing bioinformatic pipeline. There is a Rev. Med. There is an area of medical research in the science of Pharmacol. Sci. 25, 1–6 (2021).
Walker, L. C. In order to obtain evidence relating to impact on splicing, we had to use the ACMG/AMP framework. Am. J. Hum. A person named Genet. 110, 1046–1067 (2023).
Li, H. Risks of breast and ovarian cancer for women harboring pathogenic missense variants in BRCA1 and BRCA2 compared with those harboring protein truncating variants. Genet Med. 24, 119–129, was published in 1992.
Cooper, G. M. & Shendure, J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet. 12, 628–640 (2011).
Tabet, D., Parikh, V., Mali, P., Roth, F. P. & Claussnitzer, M. Scalable functional assays for the interpretation of human genetic variation. Annu. Rev. Genet. 56, 19.1–19.25 (2022).
Findlay, G. M., Boyle, E. A., Hause, R. J., Klein, J. C. & Shendure, J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513, 120–123 (2014).
Biswas, K. et al. A computational model for classification of BRCA2 variants using mouse embryonic stem cell-based functional assays. NPJ Genom. Med. 5, 52 (2020).
A group of people have identification of different kinds of cancer genes using base editing screens. Genome Biol. 22, 80 (2021).
Bayesian analysis of BRCA2 variant frequencies associated with Fanconi anemia using a VarCall model 37, JAGS programming language and the ACMG-AMP guidelines23
It is Mishra, A. P. RAD51 recruitment is dependent upon replication inducing and meiotic DNA double strand breaks. Nat. Commun. 13, 1751 started in 1992.
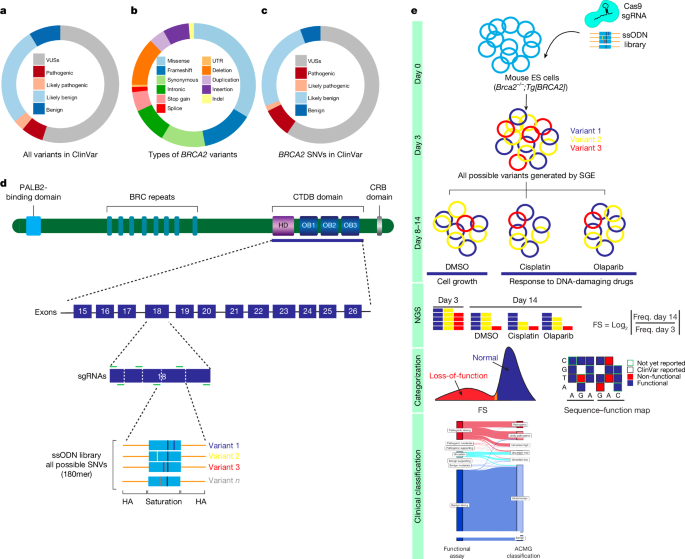
Replicate-level variant frequencies were computed at each assay time point (D0, D5 and D14) by dividing the variant read count by the replicate total for each exon. The estimated effect was made by using the ratio between the two D0 and D5 read counts. The VarCall model37 was applied to the positionally adjusted log ratio of the D14 and D0 read counts. A model with a mixture model for the variant effects can be found in VarCall. The previous analysis of the BRCA2 variant 8 has been used in the formula used here. If assumed to be and if not, each variant was assigned a indicator of its pathogenicity status. The nonsense and silent variants were thought to be benign. The measurement model adjusted for batching by including replicate by exon-level location and scale random effects and included t-distributed error terms to allow for outliers. The VarCall model was fitted using a MCMC method through the use of the JAGS language38. There were computations done in the R programming language. AlphaMissense used a previous probability of 0.2 for variant in the DNA-binding region to estimate a predicted Frequency of 0.23 for the same variant. The Bayes factor was computed using MCMC’s output. The thresholds for the Bayes factor based on strength of evidence of pathogenicity or benign level (PStrong, PModerate or PSupporting, VUS, BStrong, BModerate or BSupporting) were derived from the Bayesian interpretation of the ACMG–AMP guidelines23. The analysis details can be found in the Supplementary Methods.
S. Richards, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. It was Genet. Med. 17, 395–423.
Biswas, K. et al. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood 118, 2430–2442 (2011).
The article was titled “Ardi, M. et al.” MAVISp: multi-layered assessment of variants by structure for proteins. Preprints were published at bioRxiv.
FASTQ files of sequenced samples from Illumina MiSeq or NextSeq assays were trimmed for adapter sequences using cutadapt (v.3.5). The pair of end reads were converted into single reads. The single reads were aligned to the human reference genome (GRCh38) utilizing bwa-mem (v.0.7.17). Following alignment, the custom-developed tool CountReads was used for DNA-sequencing data analyses, with a particular focus on the identification and characterization of mutations. CountReads included the preparation of reference amino acid and DNA sequences, validation of sequencing data integrity and precise trimming of reads to relevant regions. The method was able to differentiate between different variant types and confirm the presence of specific variant data. CountReads produced a variant call format (VCF) file, which was annotated using CAVA35. The SpliceAI tool (v.1.3.1)36 was utilized to evaluate splicing effects associated with all observed SNVs.
The log2 ratio between the frequency of D14 and D0 read counts was used to measure the depletion or enrichment effect for each variant. The difference between D5 and D0 was compared for the use of a transformation. Variants with under-represented read counts (<10) at D0 and D5 were excluded from further analysis. log2 ratios of variants were linearly scaled within each exon across replicate experiments relative to median silent and median nonsense SNV values. For each variant, the average score was calculated from all non-missing values among replicates. Linear scaling was used to normalize scores across exons using median synonymous and nonsense values, similar to the within exon normalization. After completion of all data cleaning and quality control, a raw functional score was available for 6,959 SNVs (Supplementary Table 3).
BRCA2 functionally PStrong missense alterations were mapped in the DBD using PyMol software. The Protein Data Bank source file (identifier 1MJE) was downloaded from the NCBI Molecular Modeling Database. Three-dimensional structural modelling was based on the crystal structure of a BRCA2–DSS1–ssDNA complex39.
BRCA2 amino-acid sequences were obtained from Align-GVGD (http://agvgd.hci.utah.edu/). Sequence alignments were performed using ten species: Homo sapiens, Pan troglodytes, Macaca mulatta, Rattus norvegicus, Canis familiaris, Bos taurus, Monodelphis domestica, Gallus gallus, Xenopus laevis and Tetraodon nigroviridis. Sequence conservation analyses were performed on amino-acid residues that contained BRCA2 DBD functionally pathogenic variants. Align-GVGD26, AlphaMissense27, and Bayes-Del40 were used for prediction of inhomogeneity.
Other studies included a cell line–based drug immunoassay, a prime-editing-based SGE study and a mouse embryo-derived functional analysis.
The framework combines evidence from diverse sources, weighted according to strength, with each contributing source using the Moderate and strong terms (PM1, PM2, PM3). The data combine to produce variant classifications of benign, LB, pathogenic,LP and VUS9. The scoring rules were used for the classification of the cancer causing genes. The PS3/BS3 rule allowed integration of the BRCA2 functional data into the classification model. The values for functional evidence were capped at +4 and –4 on the log scale to avoid LP or LB classification with functional evidence alone. The study was approved by the Western Institutional Review Board, which exempted review of the clinical testing cohort, and by the Mayo Clinic Institutional Review Board (21-008216). The supplementary methods have detailed ACMG–AMP criteria used in them.
The LOH status of breast, ovarian, pancreatic, and prostrate cancer tumours carrying germline BRCA2DBD variants was obtained using the IMPACT datasets32. The FACETS algorithm41 was used to determine LOH from matched tumour–normal pairs. There were only samples with over 40% of the cancer content included in the analysis.
Design of a Cas9 sgRNA Co-expression construct to make individual SGS by Site Saturation Mutagenesis (SgRNA) a site-directed mutagenesis
Data shown in this paper is provided with explicit written consent of study participants, after approval from the institutional review boards.
HAP1 cells were maintained with both penicillin andstrepomycin. For haploidy sorting, 1 × 10−7 HAP1 cells were resuspended in 1 Hoechst 34580 and sorted at 4 degrees. HAP1 cells were transfected using Turbofectin 8.0 (Origene). All oligonucleotides and primers were synthesized by Integrated DNA Technologies.
Exons 15–26 are the ones that contain the BRCA2 DBD, and were selected for SGE. Exons 18 and 25 were split into amino-terminal-targeted and carboxy-terminal-targeted regions because of their large exon size, which resulted in a total of 14 SGE target regions. Benchling was used to design multiple sgRNAs. sgRNA-annealed oligonucleotides were ligated into pSpCas9(BB)-2A-Puro (PX459 v.2.0) (Addgene, 62988) following BbsI (New England Biolabs, R0539L) digestion to create a Cas9–sgRNA co-expression construct for each individual SGE. To make each SGE, 6001,000 bp homologous arms upstream and downstream of the target region were amplified by wild-type HAP1 g DNA and cloned into a BamHI-HF- Digested pUC19 vector using the NEBuilder HiFi DNA assembly cloning kit. Cloned plasmid backbones were subjected to site-saturation mutagenesis by inverse PCR34 using mutagenized codon NNN primers for all possible nucleotide changes at each amino-acid position. After successfully editing a silent sgRNA in the protospacer adjacent motif site, a site- directed mutagenesis was introduced into the target region to prevent it from being cut again. The introns of each arm had a single 3-nucleotide variant introduced to facilitate reamplification of the targeted DNA.
A machine learning-based algorithm has been used to predict the risk of breast and ovarian cancer in women with BRCA2 variants, based on genome-wide transcriptome data. A computational model was used for classification of BRCA2 variants using mouse embryonic stem cell-based functional assays. Genetic variants in BRCA2 are associated with breast cancer and ovarian cancer, respectively.