The ancient genomes of 100 countries show population turnovers
by admin
Random selection and ancestry-based frequencies of chromosomes. The case of the P1 and P2 SNPs
LDA therefore takes an expected value of 0 when haplotypes are randomly assigned at different SNPs and positive values when the ancestries of the haplotypes are correlated.
Stage 3: P1 and P2 then merged into P3 in generation 2,900. In P3, for each combination of selection in stage 2, we simulated positive, balancing and negative selection for m0. We had 4,000 people from P3 sample as the modern population after 20 generations of selection.
balancing selection was also probed at two genes. Positive selection made the mutants allle become almost the only one in P1 and P2, but the balance selection ensured that the allele reached 50% frequencies. Positive selection and balancing in P1 and P2 didn’t have an effect on the frequencies of m1 or m2 in P3 because we set strong positive selection. If m1 or m2 underwent balancing selection in P3, its frequency slightly increased; for example, if m1 underwent balancing selection in P1, it had a frequency of 25% when P3 was created, and the frequency reached around 37.5% after 20 generations of balancing selection in P3.
LDA is defined in terms of local ancestry. Let A(i,j,k) denote the probability of the kth ancestry (k = 1, …, K) at the jth SNP (j = 1, …, J) of a chromosome for the ith individual (i = 1, …, N).
rmLDAS(,jrm;X) is the abbreviation for leftbeginarray. {\int }{{\rm{gd}}(\,j)-X}^{{\rm{tg}}}{\rm{LDA}}(\,j,l)\,d{\rm{gd}}+{\int }{{\rm{gd}}(\,j)-X}^{2{\rm{gd}}(\,j)-{\rm{tg}}}{\rm{LDA}}(\,j,l)\,d{\rm{gd}},{\rm{if}}\,{\rm{gd}}(\,j) > {\rm{tg}}-X.\end{array}\right.$$
The GLIMPSE 51-based estimation of the variance for logistic regression models with low-coverage and low-qualification models. I. Data and analysis from a large ancient DNA dataset
The painting data and the first six of the first six were linear with the last six of the GWAS model. McFadden’s pseudo-R2 measure82 is widely used for estimating the variance explained by logistic regression models. If you want to know what McFadden’s pseudo-R2 is, there is a definition.
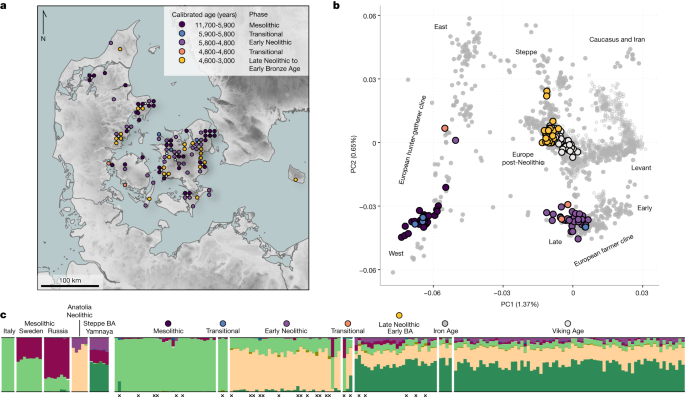
We assembled a large ancient DNA dataset to examine the variant associated with the phenotype. Here we present new genomic data from 86 ancient individuals from Medieval and post-Medieval periods from Denmark (Extended Data Fig. 2, Supplementary Note 1 and Supplementary Table 1). The samples are from the eleventh to the 18th century. We obtained ancient DNA from a tooth cementum or petrous bone and shotgun mapped the 86 genomes to a depth of genomic coverage that ranged from 0.02 to 1.6. The genomes of the 86 new individuals were imputed using 1000 Genomes phased data as a reference panel by an imputation method designed for low-coverage genomes (GLIMPSE)51, and we also imputed 1,664 ancient genomes presented in the accompanying study2. Depending on the specific data quality requirements for the downstream analyses, we used low minor allgene frequencies and low imputation quality to remove samples with poor coverage and variant sites. Our dataset of ancient individuals spans approximately 15,000 years across Eurasia (Extended Data Fig. 2).
We define the distance between SNPs l and m as the average L2 norm between ancestries at those SNPs. We compute L2 norm for ith genome.
The simulation that we did shows that our implementation protects against overfitting and searches the haplotype space effectively. This makes it a superior approach compared with HTR and GWAS to integrate SNP effects with gene–gene interactions. Its robustness is also retained when there are rare effective SNPs and haplotypes.
The effect sizes of the SNPs have been normalized by their standard deviations. We made a fixed effect size for haplotype 11XX twice as large as the average absolute effects. When rare SNPs were included, note that F_H_1=0.09
Regression with uncorrelated SNPs: a null model with overfitting in the corrected McFadden pseudo-R2 value
$${O}{i}=\mathop{\sum }\limits{c=1}^{20}{\beta }{c}{C}{{ic}}+\gamma \left(\mathop{\sum }\limits_{j=1}^{4}{\beta }{{G}{j}}{G}{{ij}}+{\beta }{{H}{1}}{H}{1}\right)+{e}_{i}+w,$$
The top M haplotypes that are chosen from forward regression should be kept. You can add another SNP to create 3M + 2 haplotypes. There are 3M haplotypes obtained by adding 0, 1 or X to the previous M haplotypes, as well as two bases of the added SNP, that is, ‘XX…X0’ and ‘XX…X1’ (as X was implicitly used in the previous step). The top M haplotypes are then selected. Repeat this process until M haplotypes are obtained that include k – 1 SNPs.
The fitted and null model have the same chances as LM and L0 do. Taking overfitting into account, we use the adjusted McFadden’s pseudo-R2 value by penalizing the number of predictors:
$${R}^{2}\left({\rm{SNPs}}\right)={R}^{2}\left({\rm{sex}}+{\rm{age}}+18{\rm{PCs}}+{\rm{SNPs}}\right)-{R}^{2}\left({\rm{sex}}+{\rm{age}}+18{\rm{PCs}}\right).$$
$${Y}{i} \sim {\rm{Binomial}}\left(1,{\pi }{i}\right){\rm{;}}\log \left(\frac{{\pi }{i}}{1-{\pi }{i}}\right)=\mathop{\sum }\limits_{k=1}^{K}{\beta }{jk}\,{X}{ijk}+\mathop{\sum }\limits_{c=1}^{{N}{c}}{\gamma }{c}{C}_{ic}.$$
$${H}_{ij}=\left{\begin{array}{ll}1, & {\rm{if}}\,i{\rm{th}}\,{\rm{individual}}\,{\rm{has}}\,{\rm{haplotype}}\,j\,{\rm{in}}\,{\rm{both}}\,{\rm{genomes}},\ \frac{1}{2}, & {\rm{if}}\,i{\rm{th}}\,{\rm{individual}}\,{\rm{has}}\,{\rm{haplotype}}\,j\,{\rm{in}}\,{\rm{one}}\,{\rm{of}}\,{\rm{the}}\,{\rm{two}}\,{\rm{genomes}},\ 0, & {\rm{otherwise}}.\end{array}\right.$$
Source: Elevated genetic risk for multiple sclerosis emerged in steppe pastoralist populations
Optimal Candidate Model Selection for a Binary Case Study using Stratified Sample Sampling and Out-of-sample Variance Estimation
Step 1: Select candidate models. The goal is to get more varied models than the ones obtained with traditional bootstrap resampling83.
Randomly sample a subset (50%) of data. Specifically, when the outcome is binary, stratified sampling is used to ensure the subset has approximately the same proportion of cases and controls as the whole dataset.
In each of the ten folds, use a different group as the test dataset and use the remaining groups as the training dataset. Then, fit all the candidate models on the training dataset and use these fitted models to compute the additional variance explained by features (out-of-sample R2) in the test dataset. Finally, select the candidate model with the highest average out-of-sample R2 as the best model.
Source: Elevated genetic risk for multiple sclerosis emerged in steppe pastoralist populations
The total variance of trait explained by genotypes, ancestry and haplotypes: application to the UK Biobank 81
Data from ref was used to provide data for MS. 4. For non-MHC SNPs, we used the ‘discovery’ SNPs with P(joined) and OR(joined) generated in the replication phase. For MHC variants, we searched the literature for the reported HLA alleles and amino acid polymorphisms (Supplementary Table 3). There were 205 SNPs that were either fine-mapped or in high LD.
We used Nc = 20 predictors in the GWAS models, including sex, age and the first 18 principal components, which are sufficient to capture most of the population structure in the UK Biobank81.
The total variance of a trait explained by genotypes (SNP values), ancestry and haplotypes (described below) is a measure of how well each captures the causal factors driving that trait. We therefore computed the variance explained for each data type in a ‘head-to-head’ comparison at either specific SNPs or SNP sets. In this section, we describe the model and covariates accounted for.
We ran a transform step as in ref. 79, centring results around the ancestral mean (that is, all ancestries) and reporting as a Z score. To obtain 95% confidence intervals, we ran an accelerated bootstrap over loci, which accounts for the skew of data to better estimate confidence intervals80.
m,effectrmpaintingrm, irmanc right
There is a way that the results can be summed into the ancestries. The sum of the kth ancestry probabilities of all the individuals in the mth cluster is said to be (P_jkm). The prevalence ofMS is defined by jm as the kth ancestry’s WAP.
The standard deviation of barpi is computed. The hypothesis H_0:barpi _jk, was also tested.
To test for gene enrichment, we formed a list of all SNPs reaching genome-wide significance (P < 5 × 10–8) and, using the R package gprofiler2 (ref. 77), converted these to a list of unique genes. We then used gost to perform an enrichment test for each Gene Ontology (GO) term, for which we used default P-value correction via the g:Profiler SCS method. This is an empirical correction based on performing random lookups of the same number of genes under the null, to control the error rate and ensure that 95% of reported categories (at P = 0.05) are correct.
Source: Elevated genetic risk for multiple sclerosis emerged in steppe pastoralist populations
Study of 100 Danish ancient genomes using shotgun sequences in a clean lab facility at the Aalborg Historiske Museum, Vestsjlland and Tjrby
Authorizations for excavating the three sites, Kirkegård, Holbæk and Tjærby, were granted, respectively, to the Aalborg Historiske Museum, the Museum Vestsjælland (previously Museet for Holbæk og Omeg) and the Kulturhistorisk Museum Randers. The current study of samples from these three sites is covered by agreements given to GeoGenetics, Globe Institute, University of Copenhagen, by the Aalborg Historiske Museum, the Museum Vestsjælland and the Kulturhistorisk Museum Randers, respectively.
The 100 ancient Dutch genomes were analysed here and contributed to the 327 shotgun-sequenced genomes. All details concerning sampling, DNA extraction, library preparation, sequencing, basic bioinformatics, authentication and dataset construction are found in ref. 3 together with all site descriptions and sample metadata. A condensed list of metainformation on the 100 Danish individuals is released here (Supplementary Data 1) together with a text summarizing the study sites and skeletons (Supplementary Note 1). In brief, laboratory work was carried out in dedicated ancient DNA cleanlab facilities (University of Copenhagen) using optimized ancient DNA methods1,77. Double-stranded blunt-end libraries were sequenced (80 bp and 100 bp single-end reads) on Illumina HiSeq 2500 and 4000 platforms. Initial shallow shotgun screening was used to identify samples with sufficient DNA preservation for deeper genomic sequencing. Of the 100 samples from Danes, 65 were from tooth cementum, 29 from petrous bones, and 6 from other bones. Sequence reads were mapped to the human reference genome and followed by estimates of genetic overage, post-mortemDNA damage, and genetic sex ID. The average number of C-to-t deamination fractions for these 100 samples is 34.9%, which is consistent with highly degraded ancient DNA. We genetically identified 67 males, 32 females and one undetermined in our dataset (Supplementary Data 1).
The data was demultiplexed using the illumina software BCL Convert. Adaptor sequences were trimmed and overlapping reads were collapsed using AdapterRemoval (v2.2.4)53. BWA (v0.7.17)54 had seeding disabled so that the human reference genome build 37 would have higher sensitivity, but only if single-end collapsed reads were mapped to it. Paired- and single-end reads for each library and lane were merged, and duplicates were marked using Picard MarkDuplicates (v2.18.26; http://picard.sourceforge.net) with a pixel distance of 12,000. Read depth and coverage were determined using samtools (v1.10)55 with all sites used in the calculation (-a). Data were then merged to the sample level and duplicates were marked again.
The main population genetics approach on which we based our inference was population-based painting (detailed below). We used other standard techniques to robustly understand population structure. ThePCA was used to look at the population structure of the dataset. We used PLINK62 to make up the imputed panel. On the basis of 1,210 ancient western Eurasian imputed genomes, the Medieval and post-Medieval samples clustered close to each other, displaying a relatively low genetic variability and situated within the genetic variability observed in the post-Bronze Age western Eurasian populations.
There were 57 genome wide-significant non- MHC SNPs downloaded for RA in Europeans70. We retrieved MHC associations separately (ref. 71; with associated ORs and P values from ref. 72). In total, we generated 51 pairs of SNPs that were either fine-mapped or in high LD.
The 1000 Genomes project has a whole-genome sequence data of 2,504 individuals from 26 world-wide populations.
Supplementary Data 4 shows the analysis of IBD sharing and mixture models using the same set of inferred genetic clusters. In brief, we used IBDseq80 to detect IBD segments, a carried out genetic clustering of the individuals using hierarchical community detection on a network of pairwise IBD-sharing similarities. IBD-based PCA was carried out in R using the eigen function on a covariance matrix of pairwise IBD sharing between the respective ancient individuals. The ancestry proportion was estimated in supervised modelling of the target individuals by using non-negative least squares.
The method DATES 44 was used for admissing time for people who were associated with the FBC. The estimated time for each individual was done separately, using both hunter-gatherers and early farmer individuals.
The predictions of eye and hair colour were made using the HIrisPlex system83. We used imputed effect allele dosages of 18 out of 24 main effect HIrisPlex variants, available for the ancient samples, to derive probabilities for brown, blue and grey/intermediate eye colour and blond, brown, black and red hair colour, following HIrisPlex formulas (see further details in Supplementary Note 2). We predicted relative ‘genetic height’ using allelic effect estimates from 310 common autosomal SNPs with robustly genome-wide significant allelic effects (P < 10−15) in a recent GWAS of height in the UK Biobank84. The iPSYCH 2012 case-cohort study62 used a sample of height to calculate polygenic score for ancient individuals and 3,467 Danes from a random population subcohort. Supplementary Note 2 has more details. The statures of a small number of the Danes were not suitable for estimation by actual measurement.
Bulk collagen isotope values of carbon (δ13C) and nitrogen (δ15N) represent protein sources consumed over several years before death, depending on the skeletal part and the age at death of the individual94,95. 13C values show the proportion of marine versus Terrestrial, whereas 15N values show the amount of trophic material that was acquired. For more discussion, see the supplementary note 4. Supplementary Data 2 and Supplementary Note 4 contain information on Stable isotope values from all 100 skeletons, and further discusses the full isotopic measurement in more detail. According to standard protocols, most of the 13C and 15N were conducted at the 14C Centre. Measured uncertainty was within the generally accepted range of ±0.2‰ (1 s.d.) All samples were within the acceptable range of C:N, which showed a low likelihood of diagenesis.
Strontium isotope analyses can provide a proxy for individual mobility102,103,104. The 87Sr/86Sr ratio in specific skeletal elements may reflect the local geological signature obtained through diet by the individual during early childhood and it will usually remain unchanged during life and after death105. Ongoing controversies exist over the exact use of geographically-defined baseline values106,107, which is why we restrict our observations and interpretations of Sr variation to patterns that are only relative to our own data. Data can be found in supplementary data 2, which was obtained by measuring the 87Sr/86Sr ratios in teeth and bones. For further details see Supplementary Note 5.
Using a high-resolution pollen diagram from Lake Højby, Northwest Zealand108, we reconstruct the changes in vegetation cover during the period 5,000–2,400 cal. bc using the landscape-reconstruction algorithm (LRA109,110). There is a low temporal resolution example in the refs. This is the first time that this method has been applied to a pollen record in a country other than the US. In total 60 pollen samples between 6,900 and 4,400 cal. The temporal resolution between samples is close to 40 years. The model used to estimate regional vegetation was based on data from six other lakes. 6.1 From this, regional pollen rain is calculated and local scale vegetation around Højby Sø calculated using the LOVE model110. For 25 wind pollinated species, average pollen productivity estimates were applied. The reconstructed cover for plant species were then combined into four land cover categories, crops (only cereals), grassland (all other herbs), secondary forest (Betula and Corylus) and primary forest (all other trees). The vegetation reconstruction from Højby Sø is used to illustrate the vegetation development at the Mesolithic/Neolithic transition in eastern Denmark. Supplementary Note 6 gives more information.
An international team of scientists has presented genomic data of 86 ancient individuals from and post-Medieval period from Denmark. The genomes of 86 newindividuals were imputed using 1000 Genomes phased data as a reference panel by an imputation method designed for low-coverage genomes. We also imputed 1,664 ancient genomes presented in the accompanying study.